* Negative sampling Rate (NR)*
*Comparison with SOTA methods (Features)*
*Random Seeds Exploration*
*models-dimensions-and-kernels*
Here we provide an extra page to store other results that are interesting but limited to the page of our manuscript.
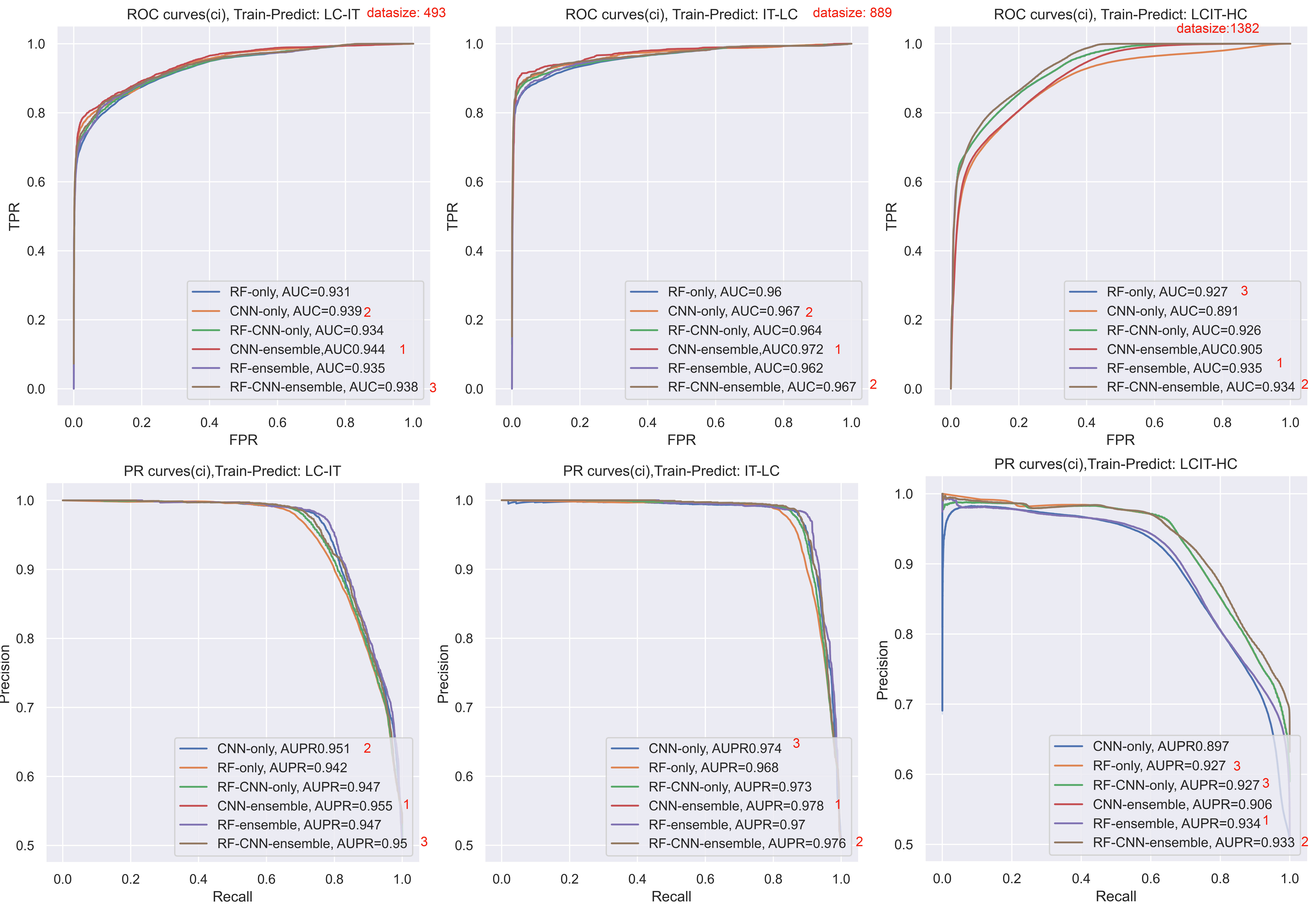
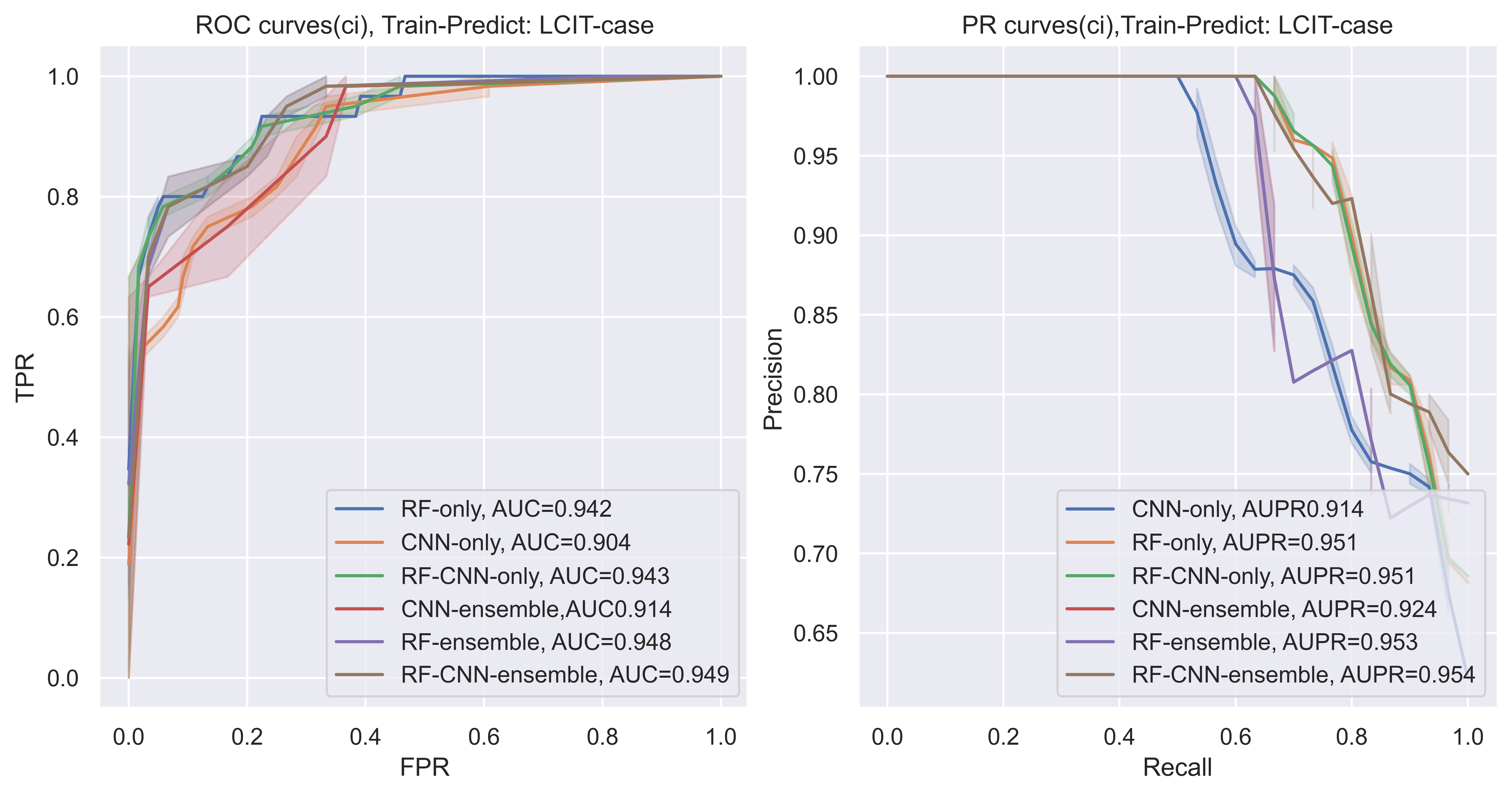
As can be seen from the picture, the performance of ensemble methods like RF-ensemble and CNN-ensemble are increased compared with their sub-model for each dataset. Also, the RF ensemble can get the highest AUC and AUPR in the third task. The RF-CNN-ensemble ranked second in this task.
the IT + LC can increase the predictive ability. Thus, in the presion@k tests,
we also leveraged the LC+IT dataset for training the models.
Thus, in this situation, the LCIT-HC is more likely to be our
situation. As, in the real scenario, the pairs to be predicted are huge and the
training dataset is limited but should be as big as possible.
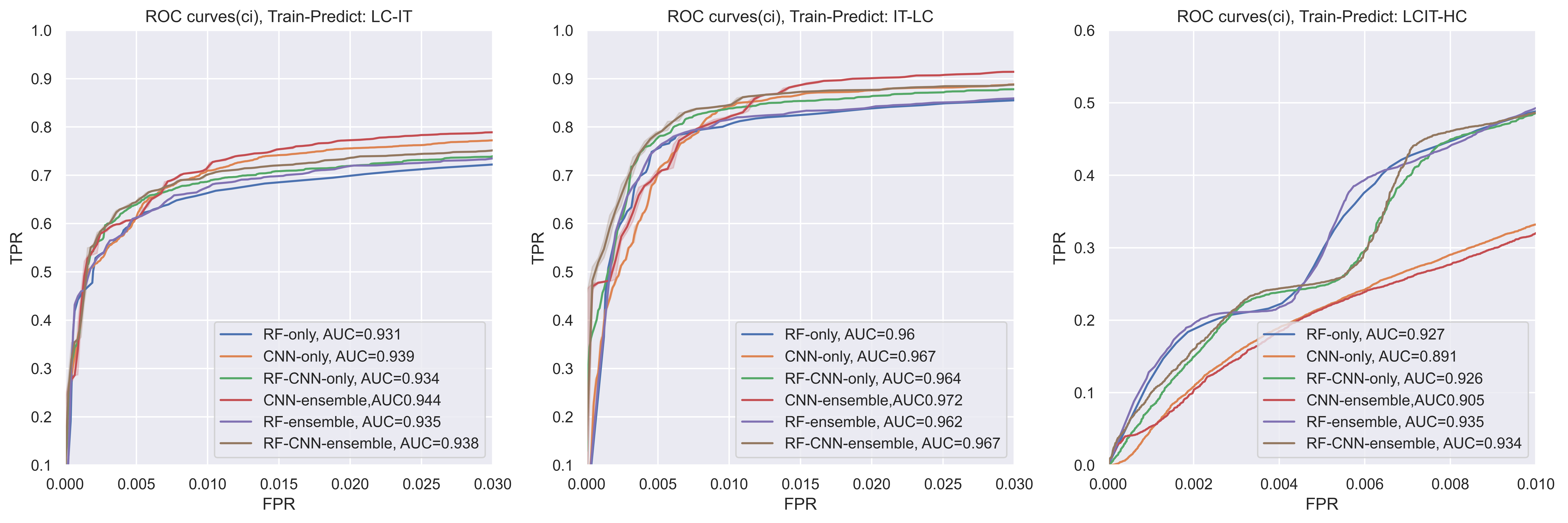
when the FPR cut into a small part (as we want to predict top related relations), we can see that for the LC dataset, when FPR is small, the RF/RF-CNN-ensemble skyrocket. For the IT dataset, before FPR is smaller than 0.01, RF-CNN-ensemble get the highest AUC. Furthermore, when it comes to big dataset, when the FPR is small, the RF and RF-CNN are always with higher TPR. which means that the predicted methods might predict more ‘True Positive’ samples when the ‘False Positive rate’ increase.
Considering we have 12417 lncRNAs and 16127 genes.
The total pair is 200,000,619, thus, even a small part of True positive rate increase, it may increase the preditive ability a lot.
that is also, maybe why, the top 100 Precision@K counts (TABLE 1) for RF-CNN-ensemble is much more than the CNN ensemble only and RF-ensemble only.
Table 1. the prediction ability of the different ensemble strategies
| Precision@K | RF ensemble (training included) | CNN ensemble (training included) | RF-CNN ensemble (training included) |
| 10 | 0.1 | 0 | 0.5 |
| 20 | 0.15 | 0.05 | 0.65 |
| 30 | 0.17 | 0.07 | 0.57 |
| 50 | 0.1 | 0.06 | 0.48 |
| 70 | 0.07 | 0.07 | 0.47 |
| 90 | 0.06 | 0.08 | 0.46 |
| 100 | 0.05 | 0.11 | 0.47 |
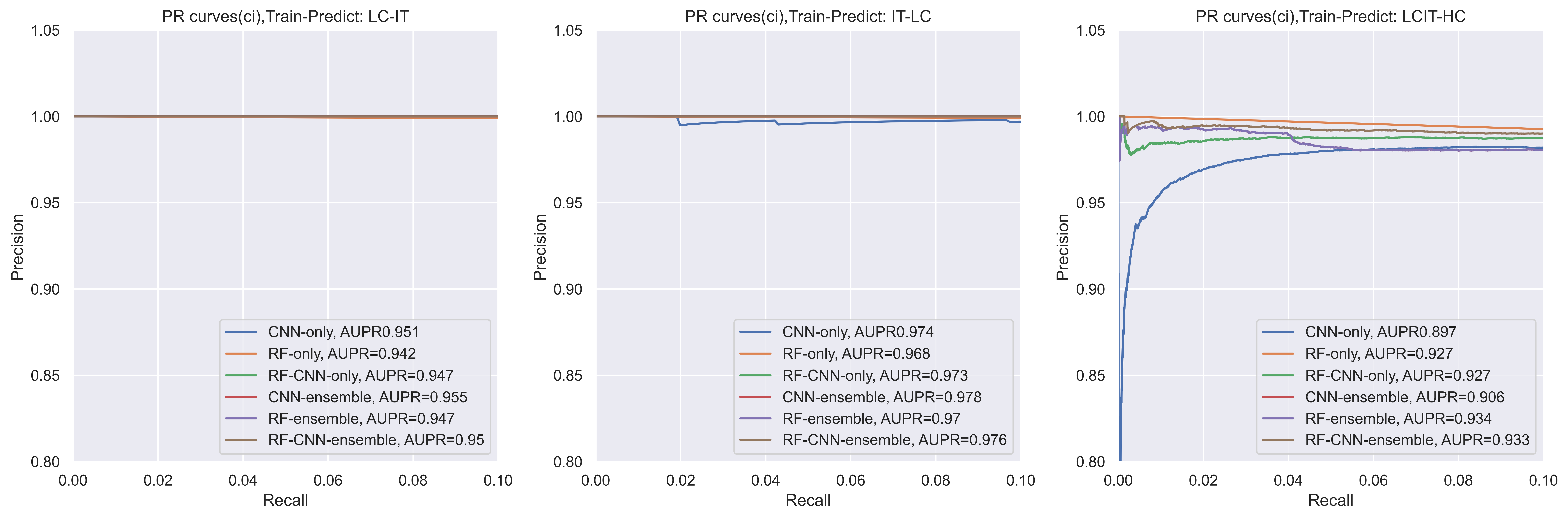
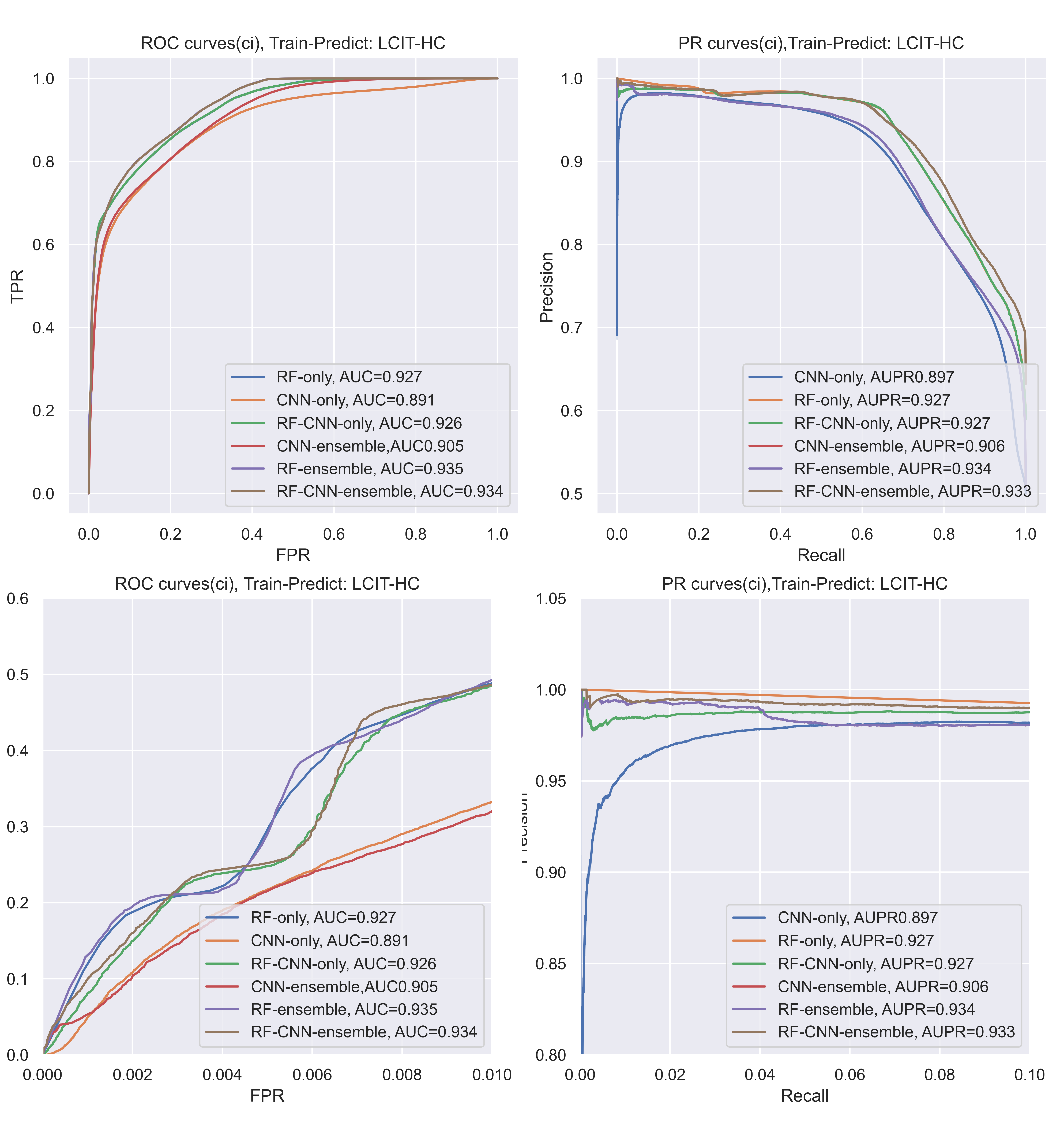
Also, if we take AUPR(PR-curves) as another example, the CNN might have a slight decrease for IT dataset when recall is small (0,02). the AUPR of CNN decreased a lot when using LCIT dataset to predict HC, thus, when added a machine learning model into the system, like the RF-CNN, it would be more smooth. If we took the ensemble methods, it would be more smooth (like RF-CNN-enmsemble).
Understanding (Details) of RF-CNN only and RF-CNN-ensemble
Take LCIT (union set) predict case as an example, we have 1-5 random seed to generate IT+LC negative samples, which contains 1382 positive samples and 1382 negative samples. Noted as LCIT_1,2,3,4,5.
Then we have 1-5 random seeds to generate the nagative samples for Case_study_6 (6 means we have 6 positive samples), which contains 6 positive samples and 6 negative samples. Noted as Case_study_6_1,2,3,4,5.
Thus,if we want to calculate the RF-only AUC, we have to concate the 12 samples* 5 (IT+LC) random seed* 5 case_study_6 random seeds
In totally, we have 300 samples. And to avoid data leakage, the ith-(LCIT)and j-th case_study_6 dataset’s score would not count, If i is equal to j, which means that if the negative sample seeds for the training set and testing set are the same, the predictive score would not be counted.
12 samples* [( 5 (IT+LC) * 5 case_study_6) -5(i=j)]=12*20=240 samples
Thus, in total, we will have 240 samples to calculate, CNN-only, RF-only, and RF-CNN only.
CNN-only 240 samples
RF-only 240 samples
RF-CNN-only,240 samples
However, for the ensemble. That is, to add all of the scores from 5 predictor( actually 4 predictor,except i=j situation) into one score. which means, for each pair in the independent dataset, it will add all of scores together to calculate the AUC/AUPR, which makes the samples to calculate the is only 12 samples *5 case_study_6 random seeds=60 samples in total.
CNN-ensemble,60 samples
RF-ensemble,60 samples
RF-CNN-ensemble,60 samples
Thus, in our Controlled deep learning methods, we actually added all predictors’ scores (3RF+3NNs) together, to satisfy the requirements for the RF-CNN-ensemble.
Other predictive results
- Top-predicted results for a selected group of lncRNA
We further selected some lncRNAs as the case study and searched the keywords pair in the PUBMED. Suppose more papers reported one pair; more possible that pair would be related to each other. We selected lncRNAs MALAT1, XIST, and DANCR, which have been widely experimented with [1], and the lncRNAs HOXA-AS2, NEAT1, TUG1, and SNHG1 according to their high rankings in the prior discussion.
The second column shows that most chosen lncRNAs’ top 20 results have more than ten pairs reported by papers from PUBMED. One exception is HOXA-AS2. The HOXA-AS2 has the fewest articles reported (74 results) among the other specified lncRNAs.
We summarized the results and regarded them as evidence to support our hypothesis (Table 2). We specifically listed the unknown target (training set removed), PMID, and published year. The mediated miRNAs are also presented. For instance, lncRNA-MALAT1 could regulate ATG in gastric cancer through miR-30e [2], exogenous lncRNA-XIST sponge miR-153 in Osteosarcoma cells, and SNAI1 was a direct messenger RNA target of miR-153 [3]. The gathered data further supported the predictive capability of the proposed approach in the context of lncRNAs acting as ceRNAs to control the target gene.
Table 2. Top unknown targets for a selected set of IncRNAs
| lncRNA | Top20 hits with possible founds | Top predicted (unknown) | PMID/ Year | miRNA involved |
| MALAT1 | 16 | ATG5 | 31926239 [2]/ 2020 | miR-30e |
| XIST | 14 | SNAI1 | 32515520 [3]/ 2020 | miR-153 |
| XIST | 14 | HMGB1 | 31418997/ 2019 | miR-29b |
| DANCR | 10 | MAPK1 | 34515297/ 2021 | miR-19a-3p |
| DANCR | 10 | CASP3 | 32581581/ 2020 | miR-758-3p |
| TUG1 | 12 | STAT3 | 34080023/ 2021 | miR‑204‑5p |
| TUG1 | 12 | DAPK1 | 30182733/ 2018 | miR-153-3p |
| TUG1 | 12 | EZR | 35012433/ 2022 | miR-377-3p |
| HOXA-AS2 | 2 | MMP9 | 29310118/ 2018 | miR-373 |
| NEAT1 | 16 | AKT1 | 33336058/ 2020 | miR-1294 |
| SNHG1 | 13 | CTNNB1 | 32239719/ 2020 | miR-4436a |
[2] Zhang Y F, Li C S, Zhou Y, et al. Propofol facilitates cisplatin sensitivity via lncRNA MALAT1/miR-30e/ATG5 axis through suppressing autophagy in gastric cancer[J]. Life sciences, 2020, 244: 117280.
[3] Wen J F, Jiang Y Q, Li C, et al. LncRNA‐XIST promotes the oxidative stress‐induced migration, invasion, and epithelial‐to‐mesenchymal transition of osteosarcoma cancer cells through miR‐153‐SNAI1 axis[J]. Cell Biology International, 2020, 44(10): 1991-2001.