The LPI-deepGBDT [1] was designed for lncRNA-protein relation prediction. The input is the RNA/protein sequence, while our input contains no protein sequence. However, we checked their GitHub repo and found that their methods provide a feature extractor. Thus, we leveraged their feature extractors for sequence and applied their features in our framework/ and GAE-LGA [2] for more evaluation.
GAE-LGA [2] , a recently published paper (27 October 2022). In their manuscript, multi-omics features for lncRNAs and PCGs were collected. Then, they fed those features into a framework that first calculated similarities between those nodes and then fed into a graph-autoencoder for potential lncRNA-PCG relations prediction.
For the comparison, as LPI-DeepGBDT provided a feature extractor function, thus we leveraged their feature extractors in our frameworks. For GAE-LGA, we replaced their features(Since GAE-LGA were using the PCG whose number is less than ours, we first had to filter the overlap between their method and ours.) with ours and LPI’s generated features and conducted their graph-autoencoder-based framework for performance evaluation.
| Methods | Year | J | Codes | Prediction type | Comparison | How (Compare) |
| LPI‑deepGBDT [1] | 2021 | BMC bioinformatics | https://github.plhhnu/LPI-deepGBDT | Protein | Yes | Their Feature/Our Framework |
| GAE-LGA [2] | 2022.10 | BIB | https://github.com/ meihonggao/GAE-LGA | PCG-Protein coding gene | Yes | Our Feature/LPI [1] feature/ Their Framework |
Part I Using different features in our framework
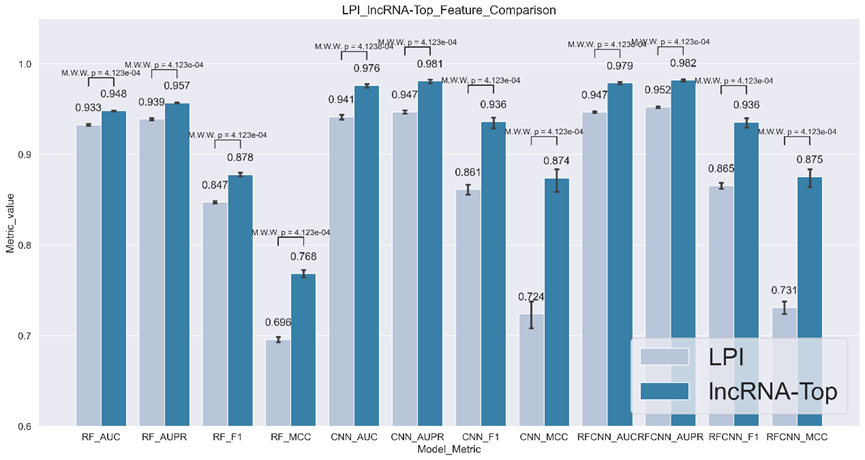
Here we leveraged the features from LPI generating features from the same group of sequences. We applied those features to the downstream framework and calculated AUC/AUPR/F1/MCC for two feature generators. Details can be found in Fig 1. As can be seen in the picture, lncRNA-top-generated features can outperform LPI feature generators in all metrics with p-value <=0.05, indicating the features are better than the SOTA methods generators.
Part II Using Features (LPI’s, ours, and multi-omics features) in the framework of GAE-LGA
Results of GAE-LGA with different features as input
| Method | D1_AUC | D1_AUPR | D1_F1-score | D1_MCC | D2_AUC | D2_AUPR | D2_F1-score | D2_MCC | D3_AUC | D3_AUPR | D3_F1-score | D3_MCC |
| GAE-LGA_ (Adjust_original features) | 0.9522 | 0.5479 | 0.7691 | 0.6071 | 0.923 | 0.5984 | 0.8265 | 0.6907 | 0.8149 | 0.4169 | 0.7761 | 0.6007 |
| GAE-LGA_(LPI_Features) | 0.9482 | 0.5407 | 0.7692 | 0.6072 | 0.918 | 0.5833 | 0.7901 | 0.633 | 0.8404 | 0.5312 | 0.8172 | 0.6524 |
| GAE-LGA_(Our_Features) | 0.9489 | 0.5453 | 0.7726 | 0.6124 | 0.9242 | 0.6176 | 0.834 | 0.7019 | 0.8568 | 0.5683 | 0.8359 | 0.6809 |
| Increment (%) | -0.346 | -0.476 | 0.4551 | 0.8730 | 0.1300 | 3.2085 | 0.9074 | 1.6215 | 5.142 | 36.315 | 7.7051 | 13.351 |
We conducted the experiments again in the adjusted network (that removed all non-overlapped lncRNA/gene and corresponding rows and columns) to acquire a benchmark value of GAE-LGA, named as GAE-LGA (Adjust_original features). Then we replaced the original mult-omics features with LPI [2] feaures and our feaures (lncRNA-Top features). The results shown that our features can outperformed most of the metrics (10 out of 12 metrics). Notably, as more lncRNA is involved in the dataset( from 117 to 155 to 193), the performance increment increases correspondingly.
Dataset Details After filtered
| Dataset | lnc | overlap_lnc | PCG (Protein coding gene) | overlap_gene |
| Dataset1 | 208 | 117 | 256 | 211 |
| Dataset2 | 238 | 155 | 716 | 617 |
| Dataset3 | 263 | 193 | 498 | 425 |
Ref:
[1] Zhou L, Wang Z, Tian X, et al. LPI-deepGBDT: a multiple-layer deep framework based on gradient boosting decision trees for lncRNA–protein interaction identification[J]. BMC bioinformatics, 2021, 22(1): 1-24.
[2] Gao M, Liu S, Qi Y, et al. GAE-LGA: integration of multi-omics data with graph autoencoders to identify lncRNA–PCG associations[J]. Briefings in Bioinformatics, 2022, 23(6): bbac452.