Controlled Deep Learning Approaches for LncRNA-Gene Relationship Annotations across Different Platforms
Introduction
Here, we designed controlled deep-learning algorithms to predict lncRNA-gene (target) regulating relations. Mechanism-based: The lncRNAs could regulate gene expression by playing a role as competitive endogenous RNAs (ce-RNAs) competing with a gene in combining with microRNA. Inspired by this mechanism, We modeled the lncRNA sequence and 3’UTR sequence of the gene. We explored negative sampling, cross-validation, four independent datasets, reliable metrics, and other details that can be omitted when conducting machine learning\deep learning on Bioinformatics. Our proposed ensemble is not only a model-based ensemble but also a random-seeds-based ensemble method. We applied Controlled Deep-Learning Approaches, which significantly increased AUC/AUPR from the transfer verification among independent datasets (compared with a single model or mode-ensemble-only results). Compared with RF-ensembles, the RF-CNN ensembles will not degenerate the AUC/AUPR a lot but can significantly increase the precision@topK. This indicated that the top-predicted pairs from lncRNA-top are more likely to be the "real-positive" pairs. The discussion shows that our model could predict top-related microRNA-mediated lncRNA targets. Even though no microRNA information is involved ( as we directly model from lncRNA to gene), as described in the manuscript, a large number of relations were related to microRNA. This probably indicates that modeled form of the mechanism is working.
Key | Features
- lncRNA-gene interactions.
- Predictive models inspired by mechanism (microRNA mediate, lncRNA play a role as ce RNA that regualte gene.)
- Gene regulatory networks
- Bioinformatics tools and analytical methods
Key | words
Controlled deep learning;
Ensemble methods;
lncRNA-gene prediction model;
—————————————————————————-
Tutorial
Before we start:
We developed software to easily access our predictive results. By now, it is only an ensembl-to-ensembl software( designed for those who are not good at coding and want to use our results), which means that the novel lncRNA is not able to be input into the system (which we will develop in the future). So far, If you input a keyword (usually gene ensemble) that is in the ensemble list, the main output window will show you it is gene or lncRNA, and if the lncRNA/gene is not in our database, it cannot give the scores. Here is a query example (tutorial) about how you can search the keywords.
Tutorial 1: Three steps to use lncRNA-top search funtion:
Step1: make sure you have these files in your folder (usually unzip the zip file should contain those files). Just check out those files before use. ‘文件’ in Chinese means file, and ‘应用程序’ means execute files. Click on the execute file, and then it start to run.
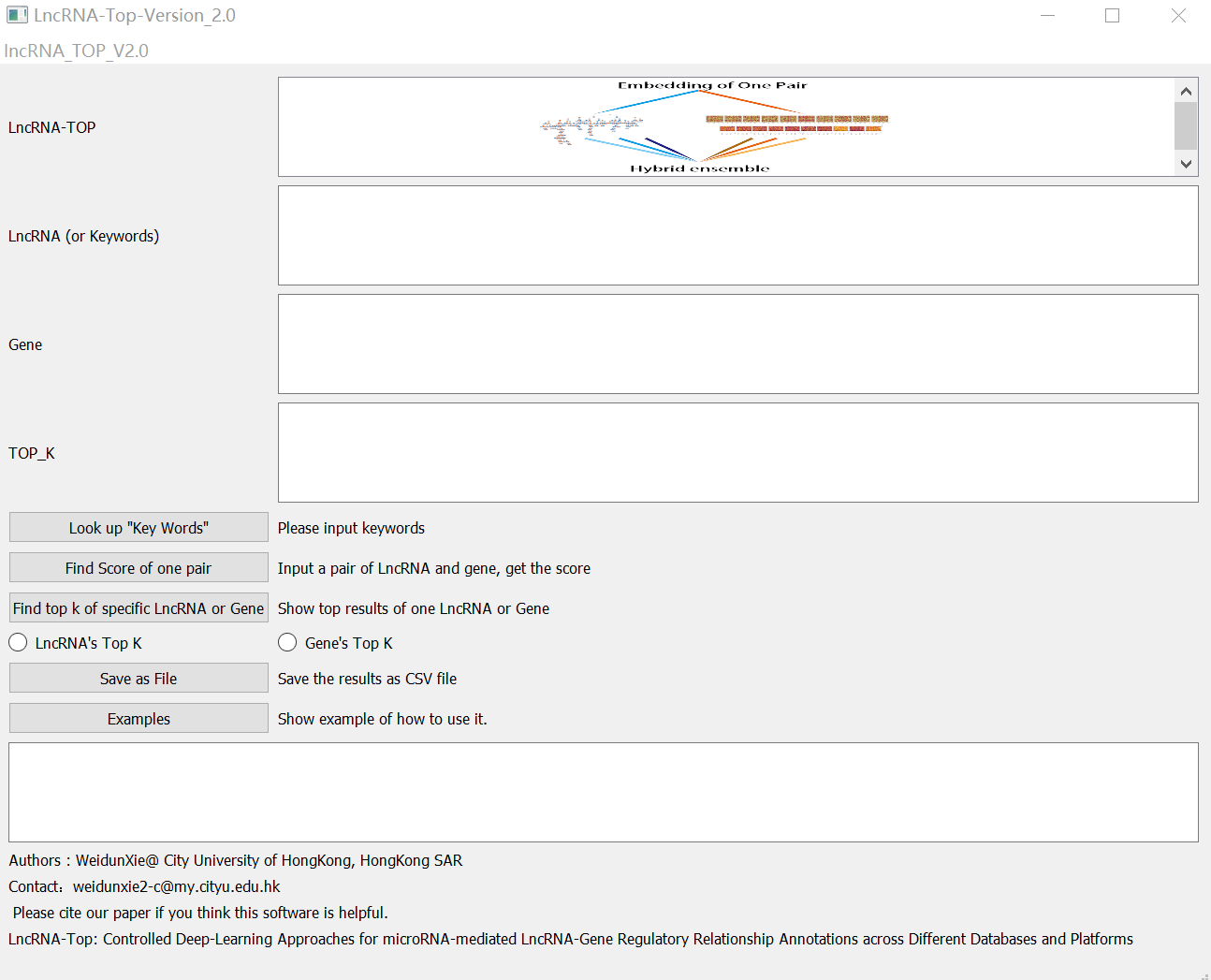
Step2: CLICK on lncRNA_top2.0.exe, you will get some interface like this.
Step3: input some ensemble, such as ‘MAT’ into the Keywords, click Look up “key words”; then you will get some results: such as lncRNA_kw: ‘lnc-ZMAT3-3’ or ‘Gene_kw:’MATN2’.
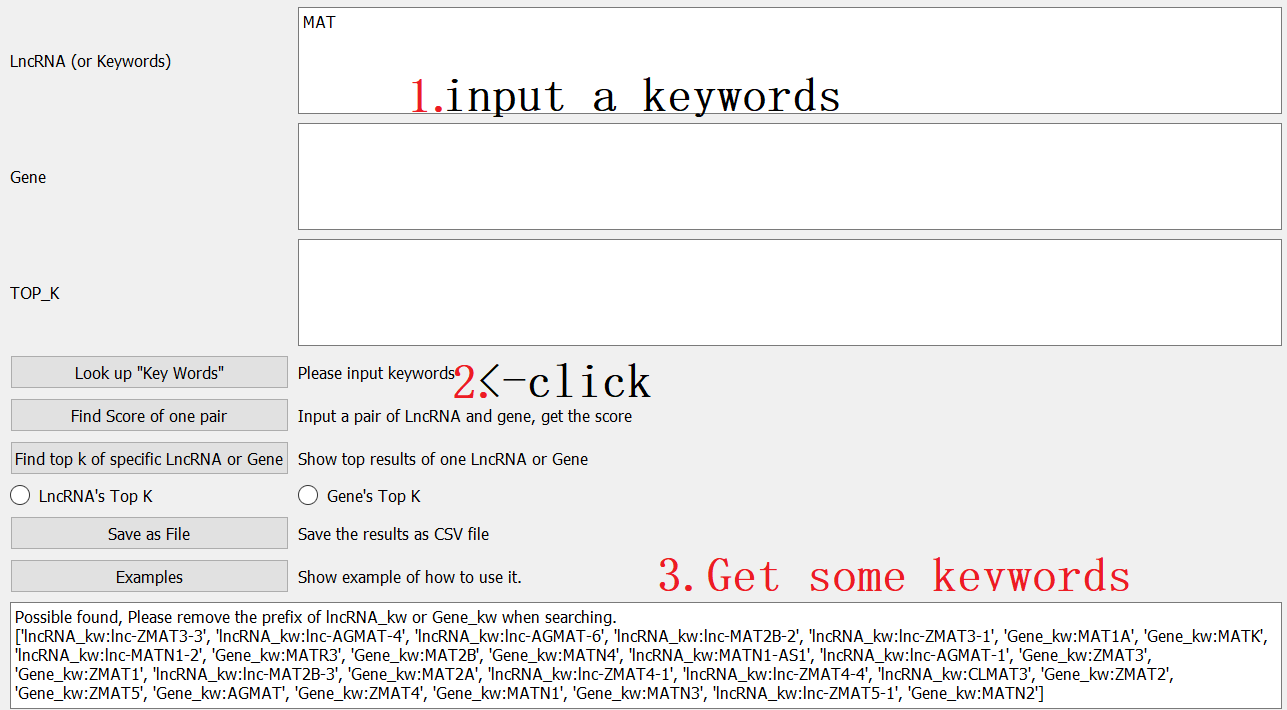
Assuming that your research is about gene MATN2, and you want to know what ‘lncRNA-top’ had predicted.
Tutorial 2: find top-predicted pairs
1) Input ‘MATN2’ into the ‘Gene’ windows, then assuming you want to get top 100 most possible related lncRNA predicted by lncRNA-top.
2) You can input ‘100’ into the ‘TOP_K’ windows.
3) And then click ‘Gene’s Top K’ botton,
4) And click “Find top k for specific lncRNA or Gene”;
5) The results are shown in the ‘Results’ windows;
6) Click ‘save as file’, then you can get the top 100 predicted lncRNAs for Gene ‘MATN2’
7) Then you can try to use the results to either construct a network, or serve as candidate targets for futher research, or as training dataset for other machine leanring tasks.
8) It is the same for ‘lncRNA’. See tutorial for ‘lncRNA’ , ‘DUBR’:
http://lncrna.cs.cityu.edu.hk/index.php/interface-software/
Our | Acknowledgments
We would like to sincerely thank all the anonymous reviewers/ editors for their valuable suggestions and advice, including:
- Report the backup server is unstable/ not responsive.
- Point out the previous version of the tutorial is not clearly written
- Point out the other issues related to our papers, including:
- Clarify more about our goal
- Clarify the input (ensemble) and output (target rank with scores)
- One of our definded metric Precision @K are lacked of definition
- Point out that we need more evidence to support our top-predicted lncRNA-gene pairs
- More experiments with SOTA methods
- Exploration with Negative Sampling Rate (NR)
- Exploration with Random Seeds.
- …
We are making progress to make it better.
About | This Server
1) Feature files
2) Dataset files
3) The interface software(for results query)
4) Source codes are also provided on GitHub: https://github.com/Xshelton/LncRNA-TOP
—————————————————————————-
About | us
Any questions/ advice, please contact:
— weidunxie2-c@my.cityu.edu.hk —
About | Our Group
visit our group websites @:
http://bioinfo.cs.cityu.edu.hk/
Back-up server@ https://lncrna.top/ (No more Support.)
About | Citations
Xie W, Chen X, Zheng Z, et al.
LncRNA-Top: Controlled deep learning approaches for lncRNA gene regulatory relationship annotations across different platforms[J].
Iscience, 2023, 26(11).